Protocols and Supplementary Data
Gilad Lab Wiki (permission required)
Protocols
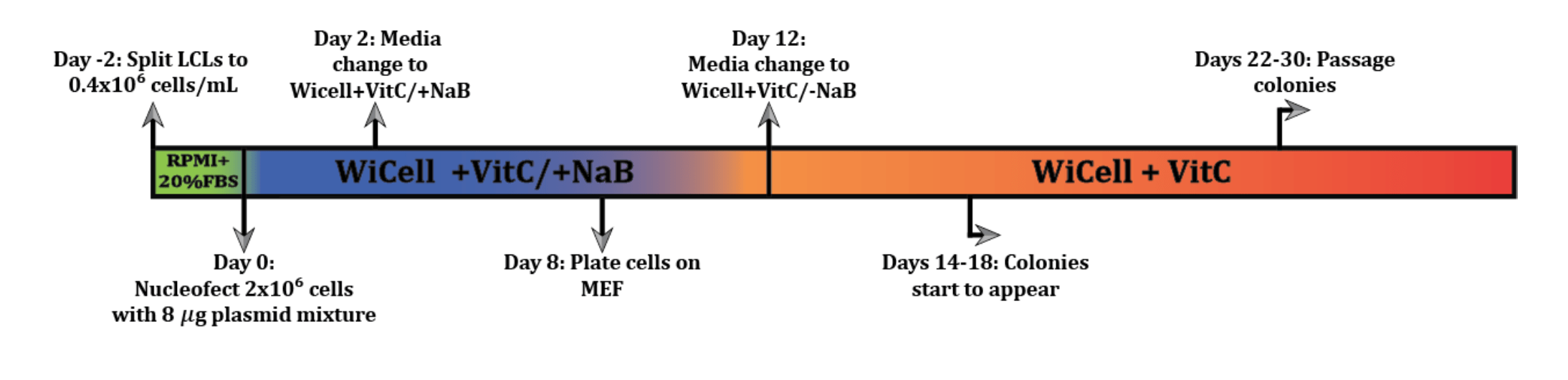
LCL Reprogramming Protocol
This protocol outlines the procedure used in the lab to generate iPSCs from LCLs.

Publications with supplementary data
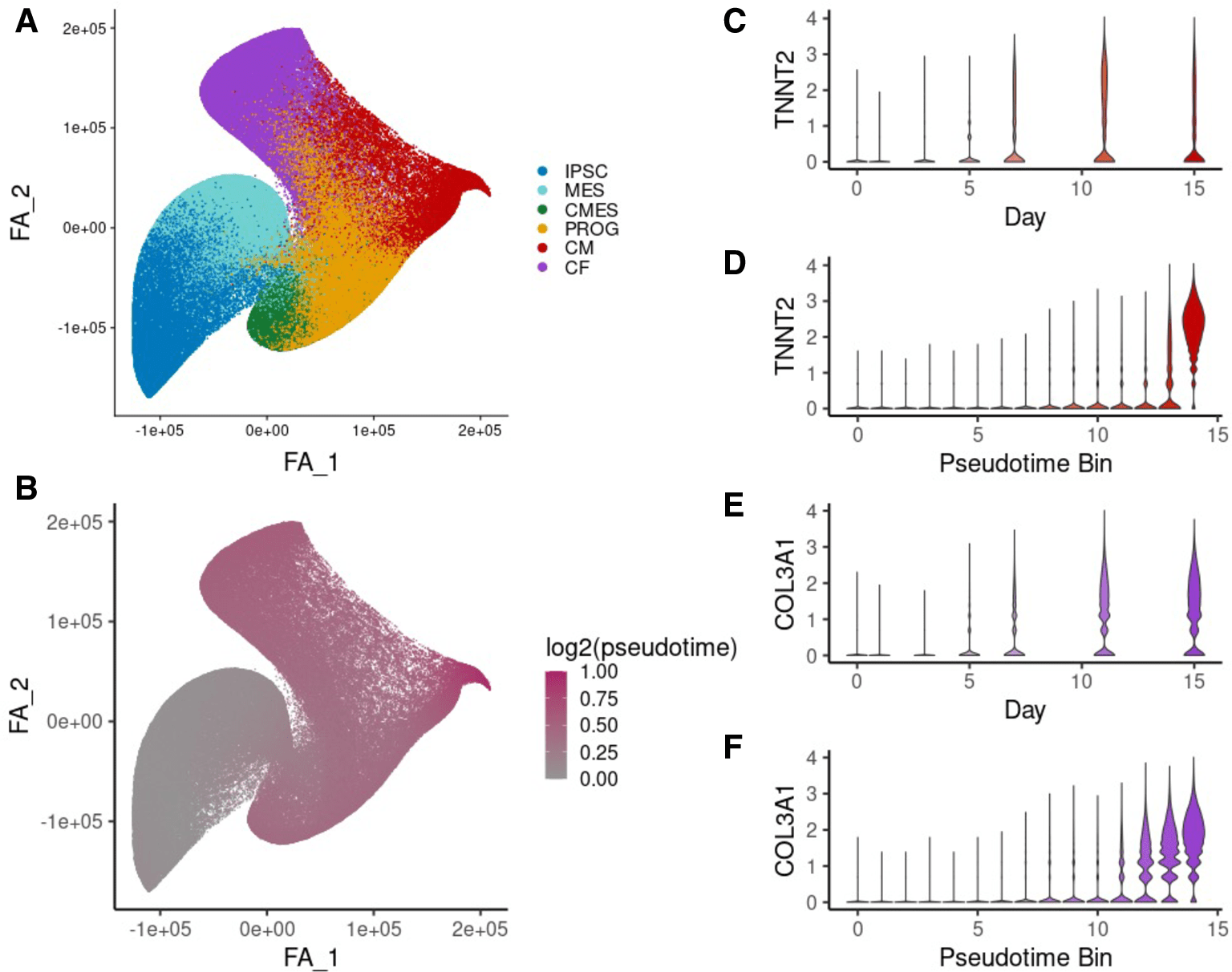
Single-cell sequencing reveals lineage-specific dynamic genetic regulation of gene expression during human cardiomyocyte differentiation
Elorbany R, Popp JM, Rhodes K, Strober BJ, Barr KA, Qi G, Gilad Y, and Battle A
PLoS Genetics; 2022
Supplementary data: upload pending
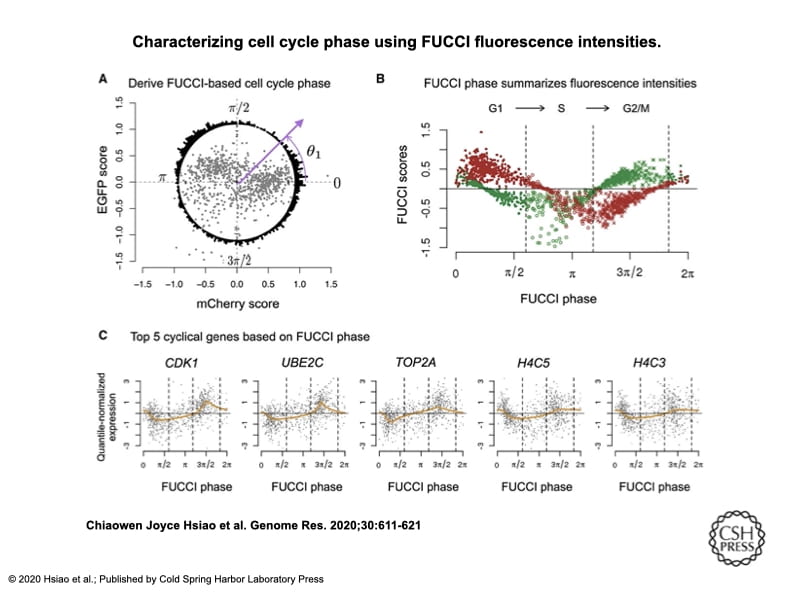
Characterizing and inferring quantitative cell cycle phase in single-cell RNA-seq data analysis
Hsiao CJ, Tung P, Blischak JD, Burnett J, Barr K, Dey KK, Stephens M, and Gilad Y
Genome Research; 2020
Supplementary data: TAR.GZ file

A comparative study of endoderm differentiation in humans and chimpanzees
Blake LE, Thomas SJ, Blischak JD, Hsiao C, Chavarria C, Myrthil M, Gilad Y, Pavlovic BJ
Genome Biology; 2018
Supplementary data: XLS file
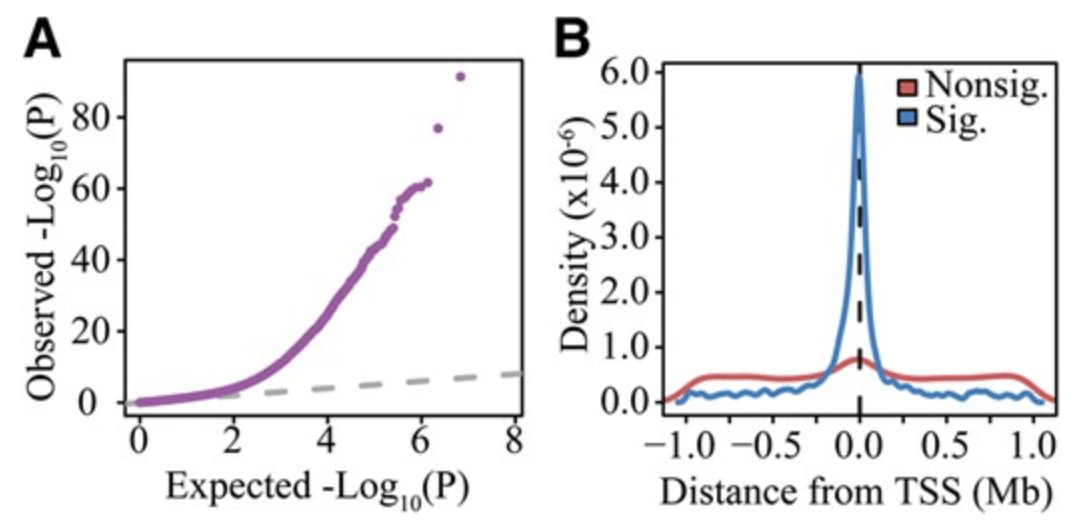
Integrated Analyses of Gene Expression and Genetic Association Studies in a Founder Population
Cusanovich DA, Caliskan M, Billstrand C, Michelini K, Chavarria C, De Leon S, Mitrano A, Lewellyn N, Elias JA, Chupp GL, Lang RM, Shah SJ, DeCara JM, Gilad Y, and Ober C
Human Molecular Genetics; 2016
Supplementary data: ZIP file
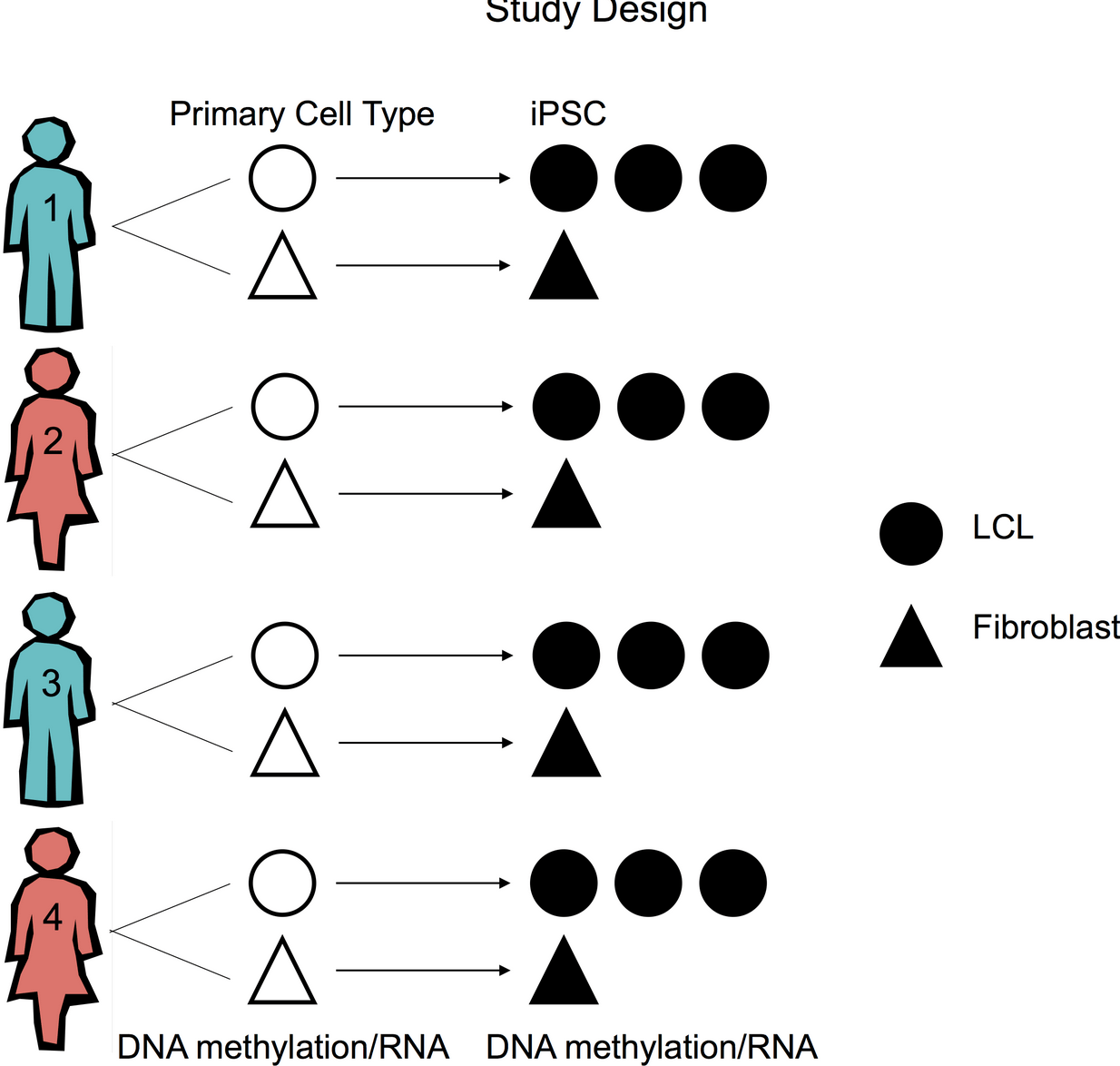
Genetic Variation, Not Cell Type of Origin, Underlies the Majority of Identifiable Regulatory Differences in iPSCs
Burrows CK, Banovich NE, Pavlovic BJ, Patterson K, Gallego Romero I, Pritchard JK, and Gilad Y
PLoS Genetics; 2016
Supplementary data: ZIP file
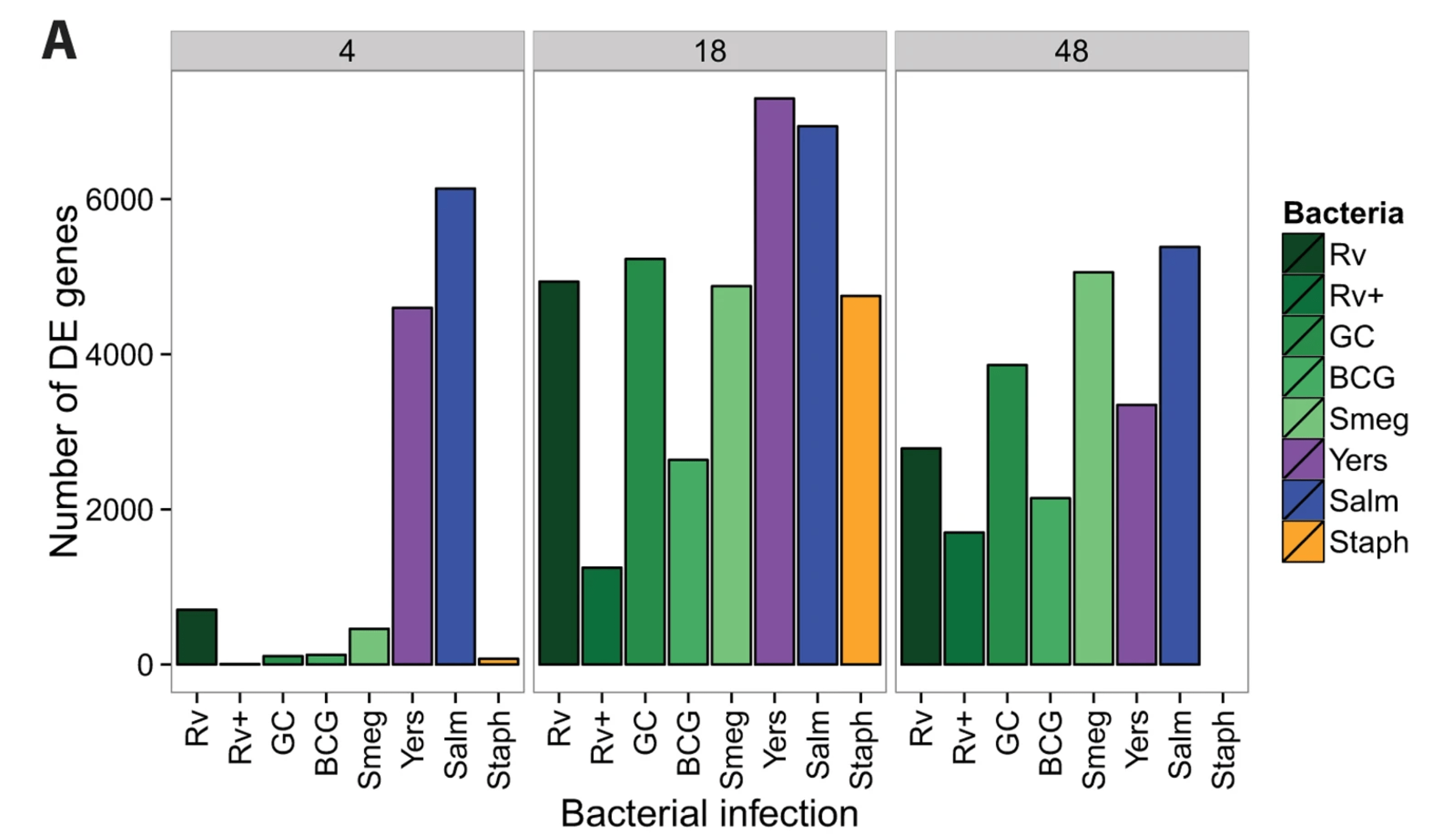
Mycobacterial infection induces a specific human innate immune response
Blischak JD, Tailleux L, Mitrano A, Barreiro L, and Gilad Y
Scientific Reports; 2015
Supplementary data: ZIP file
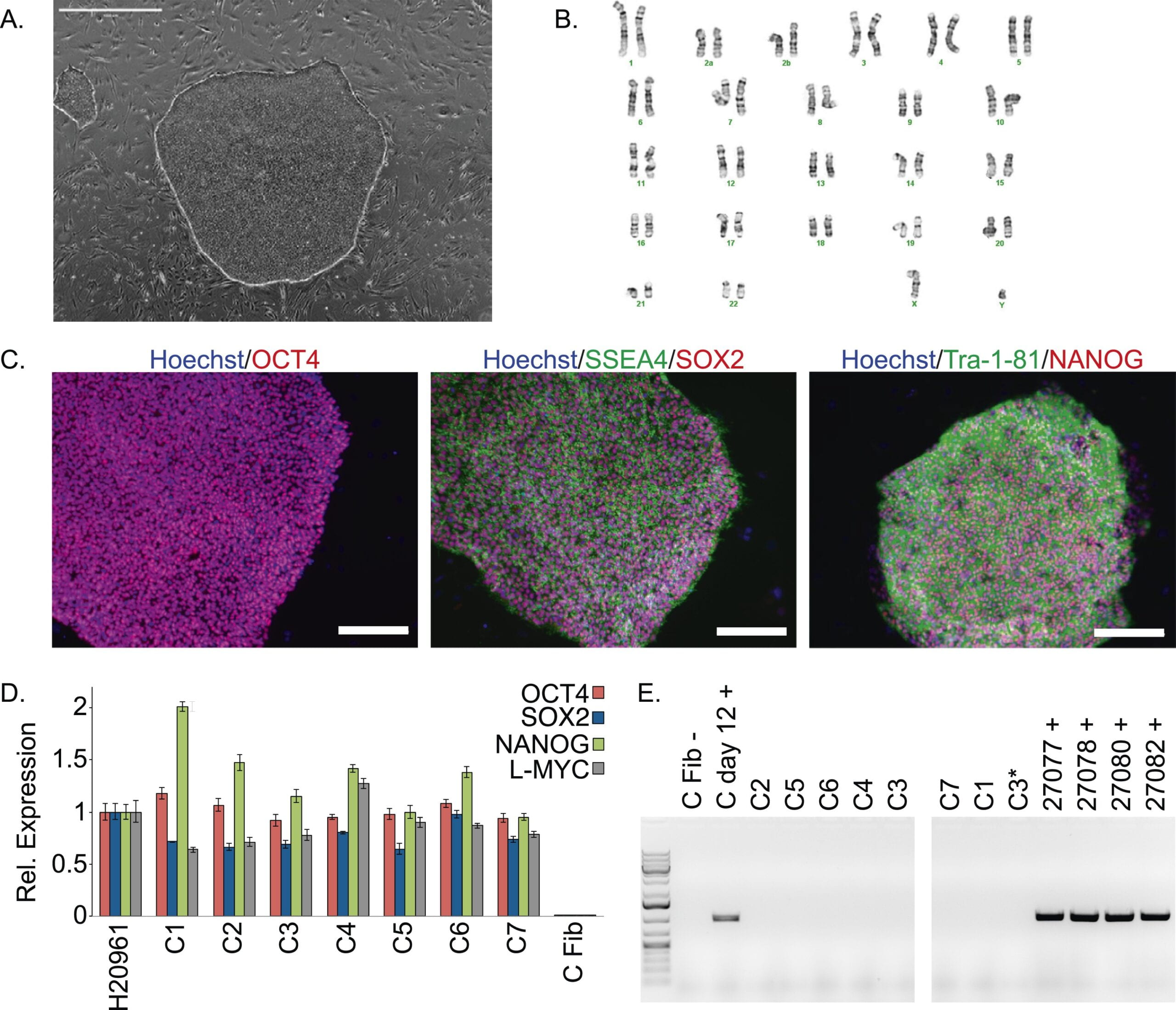
A panel of induced pluripotent stem cells from chimpanzees: a resource for comparative functional genomics
Gallego Romero I, Pavlovic BJ, Hernando-Herraez I, Zhou X, Ward MC, Banovich NE, Kagan CL, Burnett JE, Huang CH, Mitrano A, Chavarria CI, Friedrich Ben-Nun I, Li Y, Sabatini K, Leonardo TR, Parast M, Marques-Bonet T, Laurent LC, Loring JF, Gilad Y
eLife; 2015
Supplementary data: TXT file
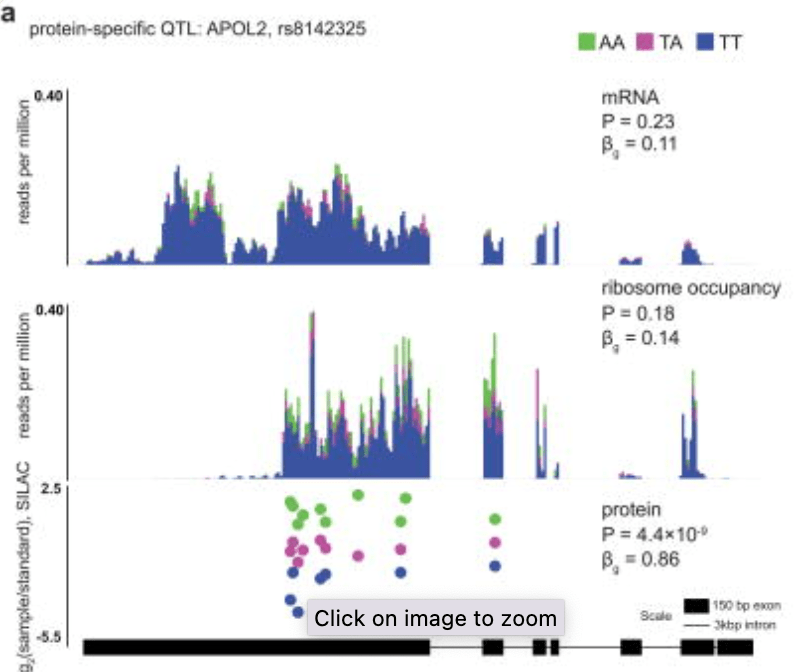
Impact of regulatory variation from RNA to protein
Battle A, Kahn Z, Wang SH, Mitrano A, Ford MJ, Pritchard JK, and Gilad Y
Science; 2015
Supplementary data: ZIP file
Genotype data and RNA sequencing data from the 72 YRI LCL lines used in this study are available on GEO (GSE19480 ) and GEUVADIS (E-GEUV-3; ArrayExpress), as is the ribosome profiling data generated in this study (GSE61742). The YRI LCLs have also been used to measure a variety of genomic phenotypes, including H3K27ac (GSE58852), DNA methylation (GSE57483), chromatin accessibility (DNAse-seq; GSE31388), transcriptional elongation rates and initiation frequencies (4sU-seq; GSE75220), RNA decay (GSE37451), and protein levels (PXD001406).
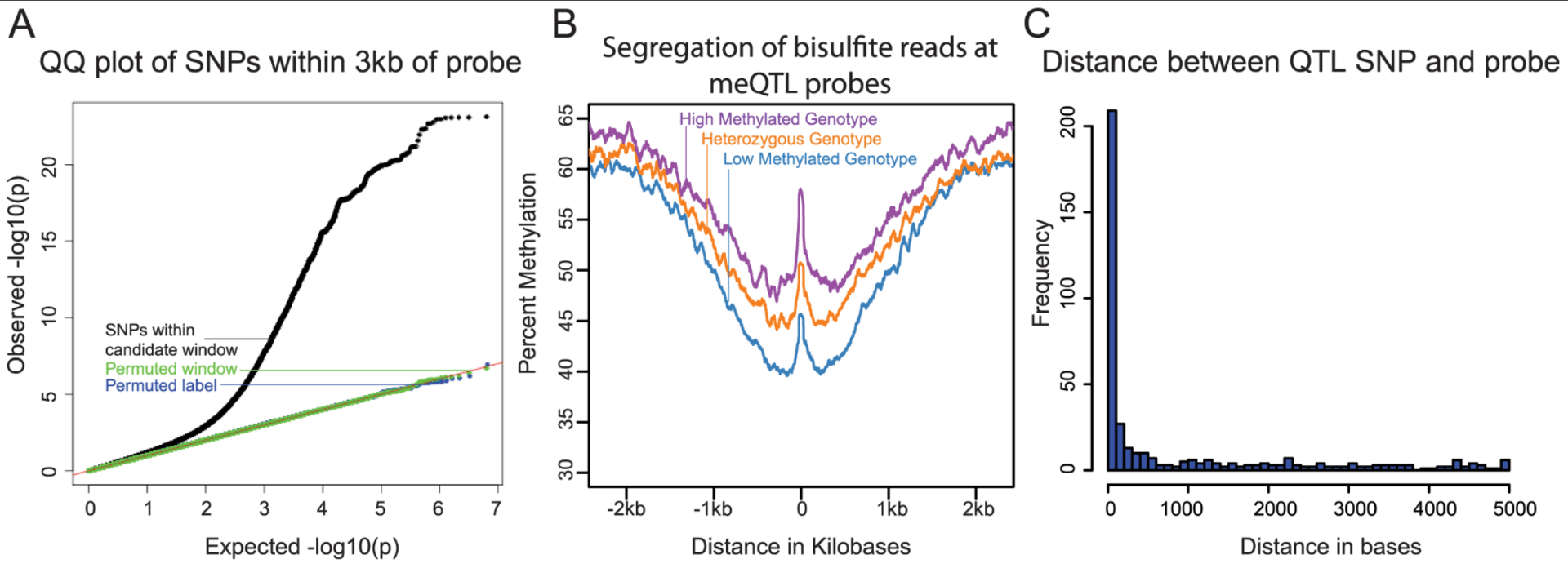
Methylation QTLs Are Associated with Coordinated Changes in Transcription Factor Binding, Histone Modifications, and Gene Expression Levels
Banovich NE, Lan X, McVicker G, van de Geijn B, Degner JF, Blischak JD, Roux J, Pritchard JK, and Gilad Y
PLoS Genetics; 2014
Supplementary data: TXT file
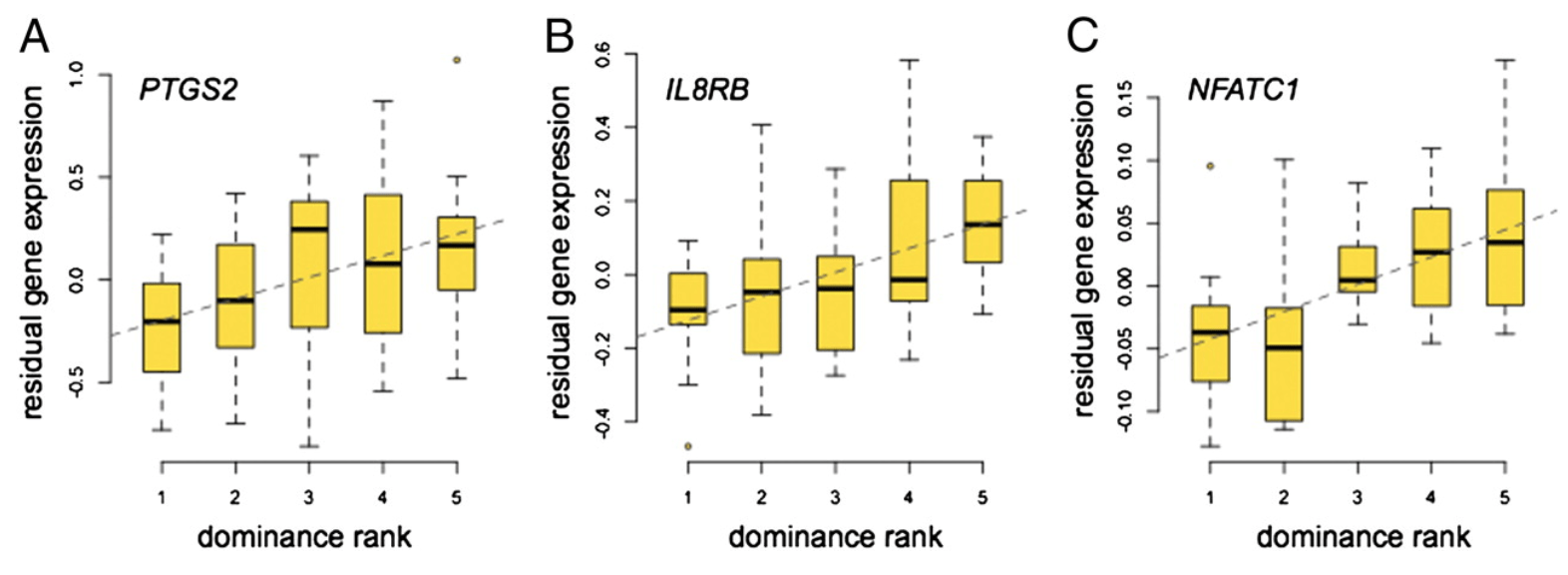
Social environment is associated with gene regulatory variation in the rhesus macaque immune system
Tung J, Barreiro LB, Johnson ZP, Hansen KD, Michopoulos V, Toufexis D, Michelini K, Wilson ME, and Gilad Y
Proceedings of the National Academy of Sciences; 2012
Supplementary data: DOC file
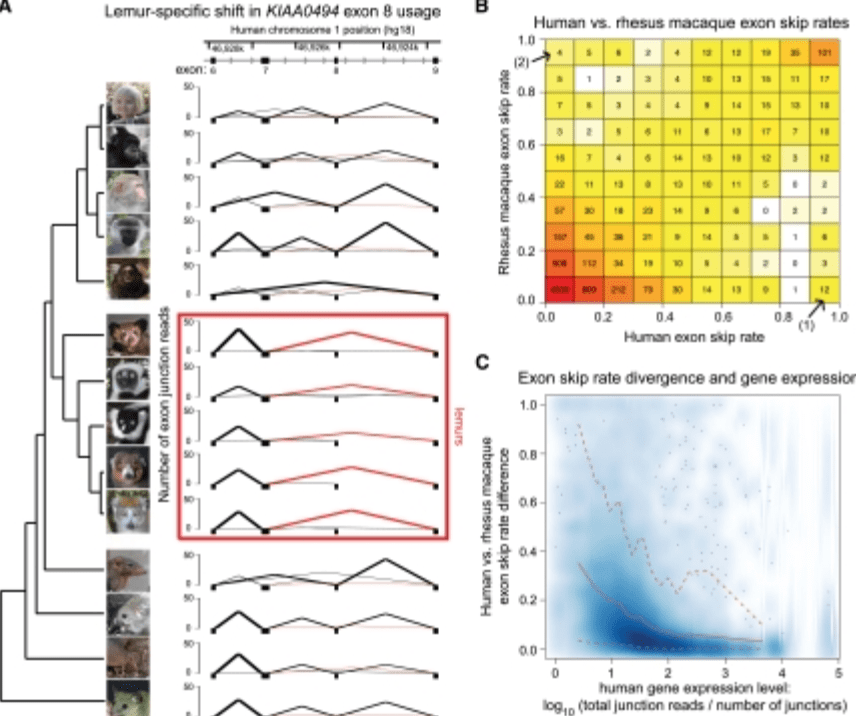
Comparative RNA sequencing reveals substantial genetic variation in endangered primates
Perry GH, Melsted P, Marioni JC, Wang Y, Bainer R, Pickrell JK, Michelini K, Zehr S, Yoder AD, Stephens M, Pritchard JK, and Gilad Y
Genome Research; 2012
Supplemental data: Please contact us if data on Genome Research webpage doesn’t meet your needs.
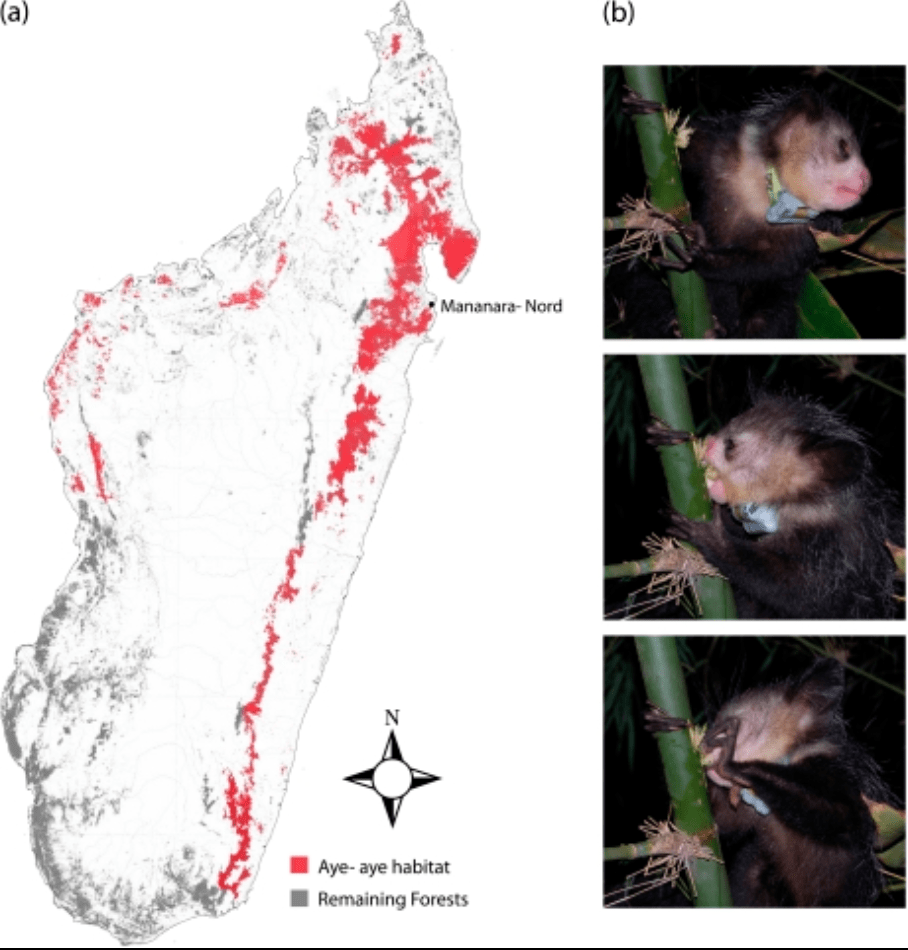
A genome sequence resource for the aye-aye (Daubentonia madagascariensis), a nocturnal lemur from Madagascar
Perry GH, Reeves D, Melsted P, Ratan A, Miller W, Michelini K, Louis EE Jr, Pritchard JK, Mason CE, and Gilad Y
Genome Biology and Evolution; 2012
Supplemental data: ZIP file
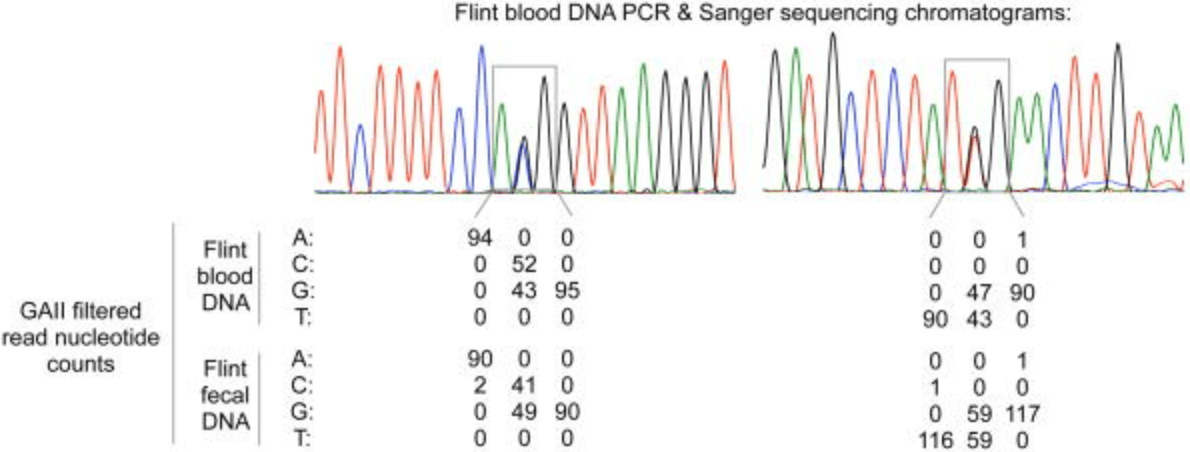
Genomic-scale capture and sequencing of endogenous DNA from feces
Perry GH, Marioni JC, Melsted P, and Gilad Y
Molecular Ecology; 2010
Supplemental data: NIH (study number SRA012374)
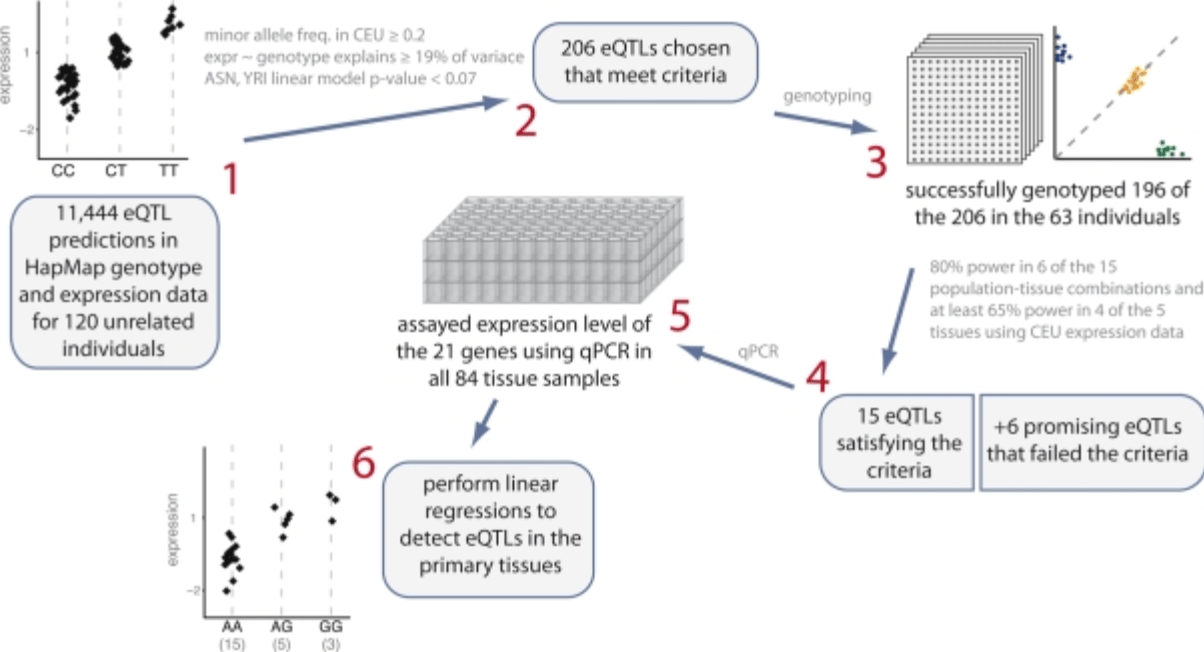
Expression quantitative trait loci detected in cell-lines are often present in primary tissues
Bullaughey K, Chavarria CI, Coop G, and Gilad Y
Human Molecular Genetics; 2009
Supplementary data: ZIP file

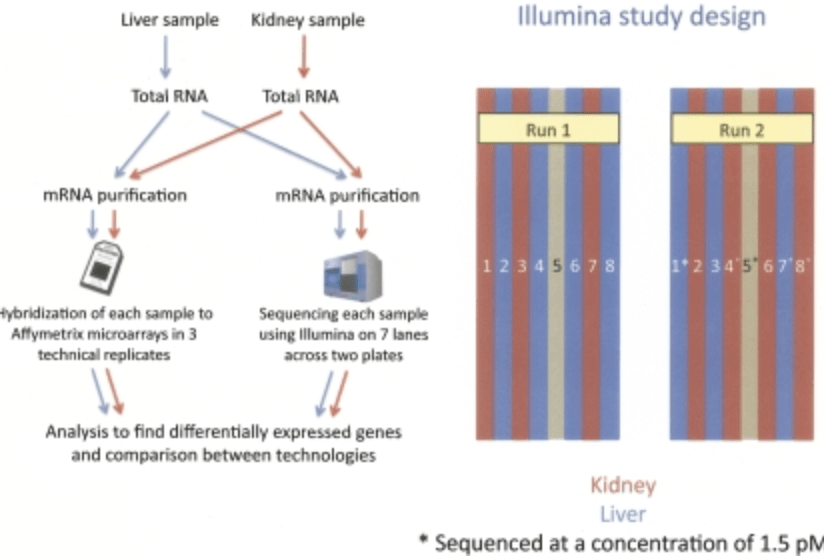
RNA-seq: An assessment of technical reproducibility and comparison with gene expression arrays
Marioni JC, Mason CE, Shrikant MM, Stephens M, and Gilad Y
Genome Research; 2008
Supplementary data: Multiple file types
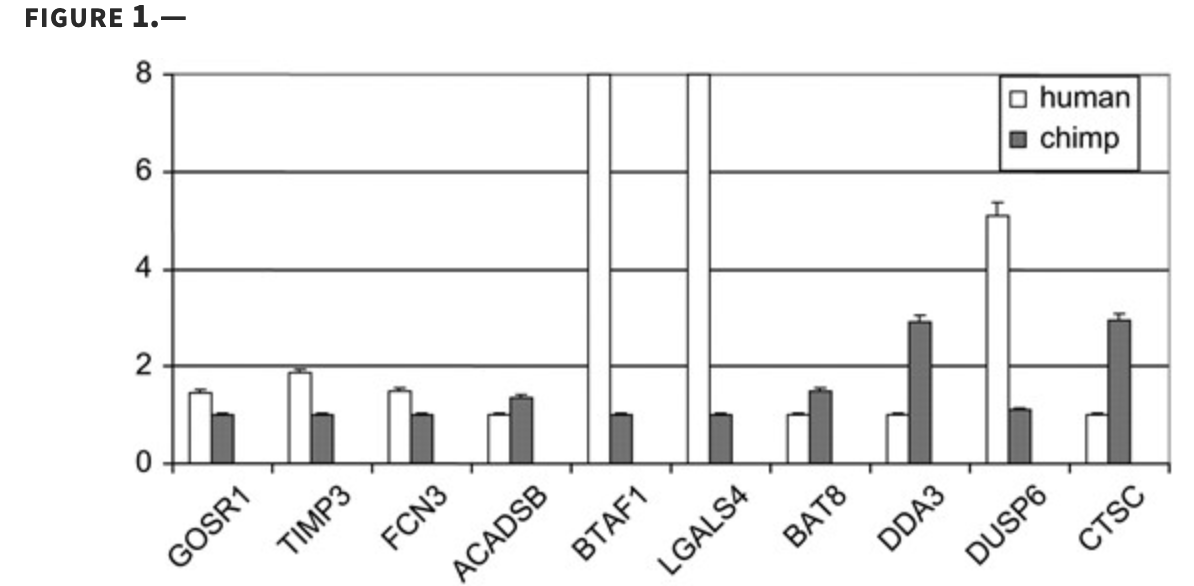
Using DNA microarrays to study gene expression in closely related species
Oshlack A, Chabot AE, Smyth GK and Gilad Y
Bioinformatics; 2007
Supplementary data: DOC file
